tRForest
Explore machine-learning predicted targets
for Transfer RNA-related fragments (tRFs)


Explore machine-learning predicted targets
for Transfer RNA-related fragments (tRFs)



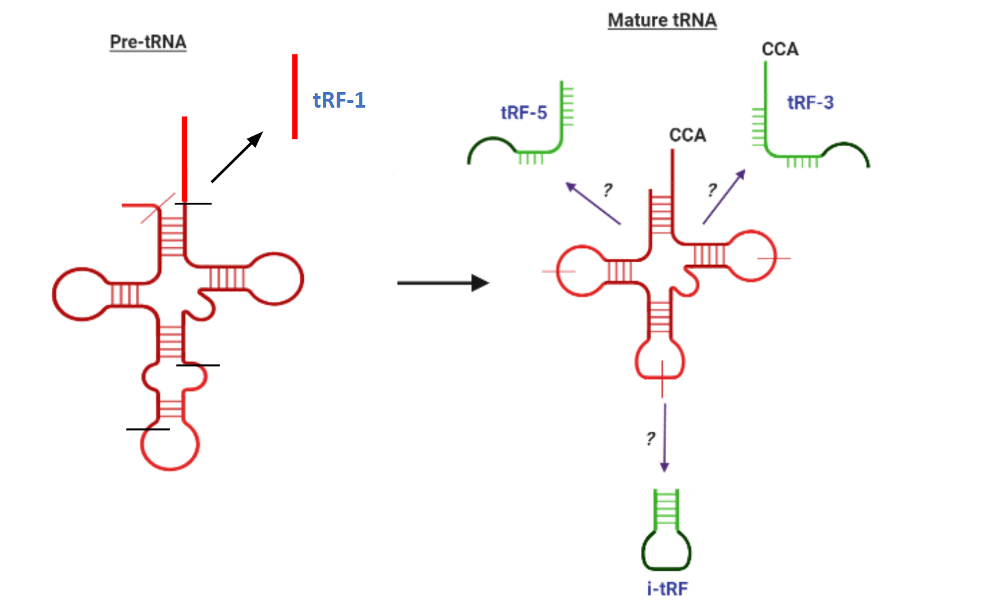
tRFs, or transfer RNA-derived fragments, are a novel class of small RNAs that are reported to associate with Argonaute proteins and post-transcriptionally regulate gene expression similarly to miRNAs. Although the mechanism of their biogenesis is unclear, they are derived from the specific cleavage of mature or precursor tRNAs. Their classifcation according to cleavage site is shown below. A complete species-wise list of known tRFs with sequence information and other information about their associated tRNAs can be found at tRFdb.
tRForest is a relational database consisting of transcript-wise tRF targets across seven species. These targets were generated using a random forests machine-learning algorithm that was trained on ground-truth tRF-mRNA duplexes obtained from a CLASH dataset of Argonaute-RNA interactions.
tRForest can be searched within a selected species by tRF-type (tRF-1, -3, or -5), tRF-ID, gene name, and Ensembl transcript ID. It provides transcript and gene information for each target ordered by prediction confidence from 0 to 1, as well as the binding energy of the tRF-target duplex, location of binding on the 3' UTR, and an interaction illustration of the duplex.
Rohan Parikh, Briana Wilson, Laine Marrah, Zhangli Su, Shekhar Saha, Pankaj Kumar, Fenix Huang, Anindya Dutta, tRForest: a novel random forest-based algorithm for tRNA-derived fragment target prediction, NAR Genomics and Bioinformatics, Volume 4, Issue 2, June 2022, lqac037, https://doi.org/10.1093/nargab/lqac037